Gene details¶
In the results panel on the right, Gravity is adding a Gene details tab.
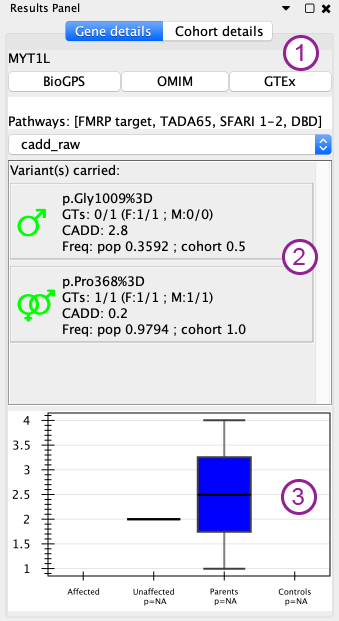
General informations (1 in figure)¶
The top of the gene details panel display the gene name and some quick links for the BioGPS, OMIM and GTEx of the gene. Additionaly the list of pathway / gene lists to which the selected gene belong are displayed in the box below.
Note
When using a GEMINI database as an input, more informations such as other names for the gene or fullname or chromosome.
Variants (2 in figure)¶
Then, there is a list of all the variants carried in the gene that passes the filter set in the control panel. This list of variants can be sorted using a variable from the dropdown list just above.
For each variants, several informations are displayed:
- a sign carrying several informations * color: same color code as for the functional impact on the network. * transmission by the mother (venus), father (mars), both (mars & venus), unknown (?), de novo (star) (alternatively in comparison mode, A, B or both to represent the carrier)
- the HGVSp annotation when available
- genotype of the individual and of his/her parents when available
- a field reserved for the deleteriousness score used (in this documentation, see the deleteriousness part in the Settings)
- frequencies in the general population and in the current cohort
A single click on a variant will copy the position (chr:position) of the variant. A double click will open in the system default web browser a configurable link for instance a web IGV with your BAM files (in this documentation, see the BAM server part in the Settings).
Enrichment (3 in figure)¶
At the bottom of this panel, there is a box plot to compare the number of variants per individual for the currently selected genes (could be multiple) accross the cohort divided into 4 categories: affected, unaffected, parents and controls. The first 3 categories are deduced from the pedigree file, the last one is optional and can be provided as a seventh column in the pedigree file (with 2 for controls, 1 for non controls, but any other value will be considered as non control).
Under the categories (other than affected) there is an indicative p-value from the Mann-Whitney U test betwwen the category and the affected. Those p-values are not corrected for multiple testing.
Note
For GEMINI database a column called Control must be added to Sample table. It must have 1 for non controls, 2 for controls any other value would be considered non controls.
